Ordering mice
Under what conditions could knockout mouse lines be provided?
The technology that was used to create the TIGM library of mutant ES cells and mice is proprietary and, therefore, TIGM operates under a license agreement with the rights to grant sublicenses to third parties. As such, please acknowledge that TIGM remains the sole owner of any materials carrying the Requested Mutation and any future transfers of such materials to other labs need to be accomplished under terms and conditions similar to those of our MTA. Also, please acknowledge TIGM in any presentations and publications reporting use of the Materials. We would also appreciate a written notice of any such publication at least 14 days prior to publication.
What are the prices for mouse lines?
Prices vary depending on the availability and source of the materials for a gene of interest, repository status, as well as if the requestor represents an academic/non-profit or commercial institution. Please contact us at info@tigm.org for a detailed price quote.
What is intended deliverable?
Each Order will be filled by the delivery of four heterozygous mice, including at least one breeding pair. Additional mice can be provided if they are available. Wild type mice from the same colony, though not necessarily littermates, can also be provided upon request. Project materials including mutation description and detailed genotyping protocol will also be provided.
Would it be possible to obtain the tissues of knockout mice before we place an order?
Most of our mouse lines exist as cryopreserved sperm, therefore tissues cannot be provided. On the other hand, we accept orders to rederive animals, and therefore we can make arrangements to supply both tissues and homozygous mice (provided that they are not embryonic lethal) of the lines produced in our facility. Please contact us at info@tigm.org for more information.
I find multiple mutations on your website for my gene of interest. Is it possible to obtain mice for each mutation?
We try to inject several clones per gene and, in most cases, only one of them produces knockout; occasionally more than one clones passes germline, and in this case we can provide both mutant lines. However, if there’s a specific need to generate two different mutants per gene, that can be arranged on a discounted basis (please contact us at info@tigm.org to discuss all of the options). If the mutations are in the same intron, it is unlikely that the phenotypes will vary between the different mice. If the mutations in the ES cell clones are in different introns, they may produce different phenotypes, each of which could be of scientific value.
Does TIGM provide subsidized lines and how do I find them in the database?
In response to RFP NIH-ES-05-04, a set of knockout mice created by Lexicon Pharmaceuticals, Inc., using targeted recombination or gene trap technology, were deposited in public repositories. Also, the Wellcome Trust has negotiated and funded the acquisition of a limited number of gene knockout mouse strains and associated phenotypic data from Lexicon. These resources have been archived and are now distributed through TIGM. A complete list of NIH and Wellcome Trust-subsidized lines is available here:[hyperlink] Data describing phenotype and gene expression in these mice were submitted to MGI and are freely available. In addition, TIGM has established a repository of over 150 selected KO mouse lines in both pure C57BL/6N and mixed 129/SvEv x C57BL/6 backgrounds: https://tigm.org/repository/. Most of these lines exist as cryopreserved sperm and will be reanimated to live mice for a significantly reduced fee for academic and non-profit researchers.
I would be interested in sending a proposal to the NIH to have the specific mouse model I am interested in added to the list of subsidized mice. Is there someone I can ask about this at the NIH?
Under NOT-DA-08-015 Lexicon made over 3,600 knockout mouse strains available for purchase by NIH funded investigators under the same terms as the contract NIH used to acquire knockout mice (RFP NIH-ES-05-04) by using NIH grant money. These mice are no longer available directly through Lexicon but are now being made available through TIGM. Also, TIGM can create novel mouse lines for an additional 10,000 genes found in its gene trap library under the same conditions. Please contact you grant administrator regarding details of immediate or future availability of such supplements
Who needs to sign the MTA?
The investigator or the laboratory PI must sign as a Recipient Scientist, and the University’s Technology Transfer Office (typically a VP or Director) must sign on behalf of the university as Customer. Your ability to quickly provide those signatures will allow us to begin the process sooner.
How long it is going to take to get the mice after we submit the MTA form?
Depending on complexity of the project, it may take between 9 and 12 months to produce mice from ES cell clones. It will take between 2 and 3 months to breed existing live mice and 4 – 5 months to resuscitate a cryopreserved line.
I queried the database but it does not produce any hits.
Don’t be discouraged by this news. Please contact us at info@tigm.org and we’ll do our best to provide you with an acceptable solution.
Can you provide information on the homozygous knockout phenotype?
We produce only heterozygous mice and ship them; therefore, we do not know the phenotype of mice homozygous for the mutation. However, the NIH-subsidized lines have phenotypic information accessible through our database. Also, for some lines viability information and some other project details (such as RT-PCR confirmation of the gene-trapping-based disruption or Southern confirmation of the targeted deletions) can be provided upon request.
Ordering ES cells
Under what conditions could ES cell clones be provided?
The technology that was used to create the TIGM library of mutant ES cells is proprietary and, therefore, TIGM operates under a license agreement with the rights to grant sublicenses to third parties. As such, please acknowledge that TIGM remains the sole owner of any materials carrying the Requested Mutation and any future transfers of such materials to other labs need to be accomplished under terms and conditions similar to those of our MTA. We also request that you return heterozygous live mice or embryos to TIGM no later than six (6) months after confirmed successful germline transmission for repository purposes. Finally, please acknowledge TIGM in any presentations and publications reporting use of the Materials. We would also appreciate a written notice of any such publication at least 14 days prior to publication.
What are the prices for ES cell clones?
Prices vary depending on the availability and source of the materials for a gene of interest as well as if the requestor represents an academic/non-profit or commercial institution. Please contact us at info@tigm.org for a detailed price quote.
What is intended deliverable?
Each Order will be filled by the delivery of two vials carrying ~5 million cells, passage number 18-19, per requested clone. Additional vials can be provided if they are available. Parental ES cell line can also be provided upon request. Project materials including mutation description, ES cell health report and detailed ES cell culture protocols will also be provided.
Are ES cell clones with homozygous mutations available?
No, the cells are heterozygous for the selected mutations (only one allele is inactivated), unless, of course, your gene is on chromosome X or Y.
Does TIGM have access to all ES cells listed in the database?
TIGM has direct and unrestricted access to all ES cell clones available in the C57BL/6 library. We also have limited access to a privately held 129/SvEv gene trap library. Some of the 129/SvEv lines listed on the site are not available to the public as they are reserved due to commercial proprietary research.
Would it be possible to obtain the cell pellets for analysis?
Yes, we can provide feeder-free ES cell pellets (both mutant clones and wild type parental cells) as a complimentary service after the full payment for ES cell clones has been received.
Why does TIGM need mice or embryos back?
TIGM has been making knockout mice for more than 4 years and we understand how difficult and expensive it is to produce and maintain live mouse lines. We ask our ES cell customers to ship the mouse lines back to us because we value each knockout line and want to ensure every single mutation is preserved for future use by the scientific community. There is no charge on your part for this service. TIGM will maintain the mice and make them available to the international community in compliance with most publishers and NIH resource sharing requirements. Should someone contact you to obtain the published mouse, you can send them to us and we will take care of the rest. These lines will be made available on a strict cost recovery basis. In addition to contributing to the scientific community, by depositing your mouse in TIGM the line will be available to you anytime in the future should you need it. Depositing your lines at TIGM also means significant cost savings to you as it allows you to eliminate the colony once your research is complete and be confident that it will be available should you decide to revisit the work.
Do I need to specify a clone when I place an order?
Our Customer Service representative can provide information on all clones available for distribution for a gene of interest and, based on that, specific clones can be ordered. But, in general, it is not necessary. You can indicate the gene you are interested in and whether you would like a true null, hypomorph or partial knockout, and our project coordinator will schedule to expand up to 4 good candidate clones. After the Quality Control is complete, you will have a chance to review a detailed report on each clone that has passed QC and after a personal consultation with our project coordinator, order those that fit your needs.
How long it is going to take to get the cells after we submit the MTA form?
It usually takes between 6 and 8 weeks to expand and QC ES cells.
Proprietary Rights
Can the ES cells, mouse and or tissues be transferred to other academic researchers?
Customer shall not transfer, by sale or otherwise, any of the Materials to any third party other than a transfer without consideration to (a) a university or non-profit entity or (b) any agency or unit of any federal, national, state, provincial, county, city or other government, domestic or foreign, in each case which is performing collaborative research with Customer involving use of the Materials. Any such permitted transfer shall be made pursuant to an agreement that includes the restrictions in Sections 3.01, 3.02, 3.03, 3.04 and 3.05 of TIGM standard MTA for the benefit of TIGM, who shall be a third-party beneficiary of the transfer agreement and shall be provided with a copy of that agreement by Customer. Please contact us at info@tigm.org for a sample of the Third Party Transfer MTA.
If I use a TIGM knockout mouse to produce a cell line, or cross-breed it to another strain, can I distribute the resulting material to other institutions?
Yes, as long as conditions specified above are met.
Concerning the fact that the distribution of a knock-out line is non-exclusive: Are the researchers having received the mice informed, if another group obtains the line?
We can inform you, upon request, if somebody else has received the line, but we will not disclose the name of the requestor.
I’m moving to another university. Can I transfer TIGM mice with me?
Such transfers are permitted as long as a third party transfer MTA is signed between the original and new institutions. Any such permitted transfer shall be made pursuant to an agreement that includes the restrictions in Sections 3.01, 3.02, 3.03, 3.04 and 3.05 of TIGM standard MTA for the benefit of TIGM, who shall be a third-party beneficiary of the transfer agreement and shall be provided with a copy of that agreement by Customer. Please contact TIGM for a sample of such MTA. . This, of course, eliminates the university you are leaving from using the line.
Technology
What do the gene-trap vectors look like?
We use different vectors to generate our ES cell clones. The actual vector design and map can be provided for each clone on an individual basis upon request. As a general guidance, please refer to the following schemas:
Gene trap vectors used in 129/SvEv library (OSTs)* 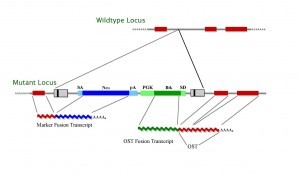
Gene trap vectors used in C57BL/6 library (ISTs)* 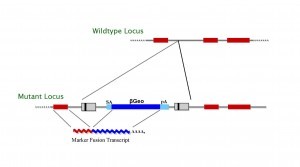
LTR, long terminal repeat; PGK, phosphoglycerate kinase-1 promoter; SD, splice donor sequence; SA, splice acceptor sequence; Neo, neomycin phosphotransferase gene; β-geo, galactosidase/neomycin phosphotransferase fusion gene; pA, polyadenylation sequence; Btk, first exon of the murine Bruton’s tyrosine kinase gene.
With the gene trap vector inserted in an intron, what is the likelihood that the gene is actually disrupted?
The likelihood of a gene being disrupted by the insertion of our vector in an intron is very high. The basic gene trap vectors we have used include a reporter gene downstream of a splice acceptor sequence. They are designed to function when inserted in an intron, to produce incorrect splicing of the target gene such that all exons downstream of the insertion site are not expressed. The gene trap cassette is inserted in a retroviral vector. Retroviruses insert as a single copy per locus, with no rearrangement of flanking sequences. They have a preference for insertions at the 5′ end of genes, often upstream of the initiator ATG, and the splice acceptor sequence we use does not appear to be bypassed by the RNA-splicing machinery. As a result, the majority of the mutations generated using our gene trap vectors are predicted to lead to null alleles. To date, the library was used to generate and analyze 1,155 mouse lines in which the vector insertion was confirmed to be within the gene of interest. Of these, 370 (32%) were not analyzed by RT-PCR due to embryonic lethality (315 lines) or perinatal lethality or reduced viability (55 lines). Of the remaining 785 lines, 706 (90%) showed no detectable wild-type transcript by RT-PCR and 79 (10%) showed drastically reduced levels of wild-type transcript. For selected lines showing reduced but detectable expression, quantitative PCR analysis has generally shown message levels of less than 5% of wild type controls. Further evidence that gene trap mutations produce null alleles is shown in Table below. In this table, we have gathered a small collection of independently published analyses of our mouse lines where the authors used methods other than RT-PCR to confirm that the allele resulted in a null mutation.
| Thioredoxin 2 (Trx-2) | No protein detected by Western blot | Nonn et al. 2003 |
| Mdm4 (Mdmx) | No protein detected by Western blot | Migliorini et al. 2002 |
| Insulin-degrading enzyme (IDE) | No protein detected by Western blot | Farris et al. 2003 |
| Insulin-degrading enzyme (IDE) | No message detected by RT-PCR | Miller et al. 2003 |
| No protein detected by Western blot | ||
| No IDE enzymatic activity detected | ||
| tRNA synthetase p38 subunit | No message detected by Northern blot | Kim et al. 2002 |
| No protein detected by Western blot | ||
| Wave1 | No protein detected by Western blot | Dahl et al. 2003 |
| Testis–brain RNA-binding protein (TB-RBP) | No protein detected by Western blot | Chennathukuzhi et al. 2003 |
| Insulin-like growth factor II mRNA-binding protein 1 (IMP1) | No message detected by RT-PCR | Hansen et al. 2004 |
| No message detected after extended exposures of whole-mount in situ hybridizations | ||
| Calpain 3 (p94) | No message detected by RT-PCR | Kramerova et al. 2004 |
| No protein detected by Western blot | ||
| GYS1 glycogen synthase | No protein detected by Western blot | Pederson et al. 2004 |
| No GYS1 enzymatic activity detected | ||
| No glycogen detected | ||
| Tektin-t | No message detected by Northern blot | Tanaka et al. 2004 |
| No protein detected by Western blot | ||
| Phosphatidylinositol 5-phosphate 4-kinase | No message detected by RT-PCR | Lamia et al. 2004 |
| No protein detected by Western blot |
Taken together, this data demonstrates that intragenic insertions efficiently disrupt gene transcription in vivo. Please refer to our Technology Disclosure and the following papers for more information: Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003 Nov and Hansen GM, et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008 Oct;18(10):1670-9.
Does the retroviral insertion lead to a frame shift?
No. The vector contains a splice acceptor. Insertion of such a vector into an intron leads to incorrect splicing of the target gene in a way that all exons downstream of the insertion site are not expressed. For more information about our gene trap technology please review our Technology Disclosure and the following papers: Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003 Nov and Hansen GM, et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008 Oct;18(10):1670-9.
I heard somebody warning that the inserted splice acceptor may not be able to completely inhibit usage of the wild type splice acceptor.
We do not believe that this is true. If this were the case, the wild type transcript would also be present in homozygotes for the trapped allele. Analysis of 741 non-embryonic lethal mouse lines demonstrates that gene-trap insertions within both exons and introns of the gene of interest lead to the disruption of the endogenous mRNA transcript in all cases. Of these, >96% show complete absence of WT message, with the remaining lines showing an average reduction in mRNA levels of 91.6% as measured by quantitative PCR (Zambrowicz et al, 2003). Also, our studies indicate that all alternatively spliced transcripts were successfully trapped in our gene trap mutants (Hansen GM, et al., 2008) These data demonstrate that intragenic insertion efficiently disrupts gene transcription in vivo and can be used to reliably predict mutagenicity before mouse production.
The insertion of the trapping vector is in an intron upstream of the ATG. Does it result in a knockout?
Our vectors are designed to disrupt a transcript and prevent its translation, even if they are inserted upstream of the ATG. For more information please see: Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003 Nov
What are the odds of producing an incomplete knockout or a hypomorph from the TIGM ES cell clones?
We always try to choose the best clone available with the gene trap inserted as close to the 5’ end as possible, to ensure the complete disruption of the gene. However, in some cases, the only clone available may have an insertion near the middle of the gene, or close to the 3’ end. Such insertions are not desirable and may produce functional isomorphs. If this is the only clone available, our TIGM scientists will review the situation with the prospective buyer and ultimately let the researcher decide whether he/she wants to generate such a knock-out, before we enter into a formal agreement. By the same token, many genes contain alternative transcription start sites, and we will try our best to select the clone that will disrupt all transcripts. However, it may not always be possible; in such situations, we will present our findings to the prospective purchaser and let him or her decide whether they want to generate mice from the available clone, before we enter into a formal agreement.
If my gene is trapped in both C57BL/6 and 129/SvEv cells, which one should I use? Apart from the genetic background of the ES cells, how do they differ?
The most significant difference will be the price of ES cells, though the cost to produce animals will be the same. Also,70% of the clones in C57BL/6 background contain a lacZ-neo fusion gene cassette (beta-geo) that enables the expression of the trapped gene to be determined by X-gal staining. The choice of one over the other may also be influenced by the location of the insert within the gene of interest.
What are germline transmission rates for the C57BL/6 and 129/SvEv clones?
Based on our experience from 100 C57BL/6 clone projects as quality control for our gene trap library, we should expect a germline transmission rate between 40-50%, when single clones are microinjected twice into host blastocysts. At the same time, 129/SvEv clones usually show a better rate of 70%. Keep in mind, though, that our staff has a lot of experience with our proprietary ES cells and that the actual germline transmission rate may be even lower in the hands of researchers not associated with TIGM. Therefore, we strongly recommend that you consider contracting TIGM to produce chimeras or heterozygous mice for you.
When I search the TIGM database, I find clones available for my gene, but when I discuss their purchase with TIGM scientists, they indicate that they are not suitable for mouse production. Why?
There could be several reasons:
- Sometimes the database may show a hit. However, when you click on the UCSC browser link, you won’t see a gene trap insertion present in the gene of interest. The false hit is sometimes generated when there is an insertion in a neighboring gene.
- Sometimes the clones with inserts in your gene of interest are in the wrong orientation. A mutagenic insertion in 129/SvEv clone must have the same orientation of the OST strand with that of the target gene; a mutagenic insertion in C57BL/6 clone must have the opposite strand orientation of the forward IST relative to that of the target gene, or the same orientation in case of the reverse sequence tag. Otherwise, they will not be able to disrupt the translation of the gene. Our database has been generated automatically, it contains >10,000 genes and the algorithm used does not always allow discrimination of correct and incorrect clones.
- Sometimes the gene trap vector is inserted close to the 3’UTR. This may produce a hypomorph, which may be less desirable than a complete gene inactivation. TIGM scientists are available to discuss the possible implications of producing a hypomorph from the clone, rather than a complete gene ablation.
Could I have the schematic representation and sequence of the insert location with the trapping vector sequence and the expected transcripts, if any?
Yes, this information can be provided. Please contact us at info@tigm.org and indicate the name of the gene or clone and we will return with this information as quickly as possible. Also, once we confirm the insertion point and the integrity of the trap during our QC procedures, we will be able to provide you with a detailed mutation description that will include gene trap map, vector sequence and genotyping strategies.
Can I get a conditional mutant?
We have capabilities to generate conditional mutations via homologous recombination. Please discuss your interests with our TIGM scientists.
Can I use the heterozygous gene-trapped mice as a haploinsufficiency model?
Using gene trap alleles as models of haploinsufficiency is perfectly valid.
Do you have data based on your previous experience working with gene trapped mice?
Nearly 1,800 gene trap mice have been generated using 129/SvEv and C57BL/6 clones and all of these are available for purchase;. A list of references is available here: https://tigm.org/publications/ .
Quality Assurance and Guarantees
What are TIGM’s QC procedures?
ES cell Quality Control includes assessment of viability and morphology, insertion site confirmation, tests for multiple insertions, tests for Y chromosome loss, and tests for mycoplasma, bacterial and fungal contaminations. We will thaw out and expand up to 6 clones (129/SvEv), or up to 4 in case of C57BL/6, that carry mutagenic gene trap insertions in your gene of interest. Only the clones that are viable and contain less than 10% of differentiated colonies will be selected for further analysis. Insertion site will be confirmed via iPCR followed by sequencing of the product. This test will also indicate if a clone is mixed with other cell populations carrying different insertions. Taqman-based Neo assay will indicate the presence of more than one gene trap copy per genome as well as whether the clone is mixed with wild type cells, while Taqman-based SRY test will indicate presence of the Y chromosome. We will also perform PCR-based Mycoplasma and MLV tests of both media and DNA samples, as well as 3-week-long incubation of culture media samples with thioglycollate for the presence of various bacterial and fungal contaminations.
Why are you not sure about the insertion site? Do I have to pay the initiation fee to find it out? What if the clone is not suitable to produce a knockout?
The gene trap sequence tag for a clone is generated via an automated process. The process may produce an incomplete readout, and therefore, may not show the precise location. In order to confirm this, we will need to thaw this clone, extract DNA and perform several rounds of manual sequencing. Only then will we be able to tell you the exact location with 100% confidence. We’ll also need to make sure that this is the only insert and another copy of the gene trap is not inserted somewhere else. These steps are part of our Quality Control process. We do not want to produce mice from the clone before we double check that this is what the investigator needs. There are no extra charges for this service. TIGM will invoice the first payment only after we have the QC data and it shows that the clone meets the QC criteria.
Does a TIGM mouse come with any assurances that the gene trap event has created a null allele?
As part of our initial consultation with investigators, we try to identify cell lines with insertions near the 5’ end of genes that have the greatest likelihood of generating a null allele. Furthermore, in a study of 741 129/SvEv-generated mice (Zambrowicz et al, 2003), analysis of mRNA expression by RT-PCR in tissues known to express the trapped gene indicated that no mRNA was detectable in 96% of the trapped genes analyzed. For the others, mRNA levels were reduced by greater than 90%. In the very unlikely event of the full length transcript being expressed at normal levels in your mice, we will do everything possible to correct this problem, even if it means that we have to go back and start the project over from injection of a new clone.
Do you have data (such as 50% reduction in expression in hets) showing that the expression of the gene is indeed reduced before I purchase the mouse?
Most of the cryopreserved mice produced from 129/SvEv gene trapped ES cells have been confirmed by RT-PCR for complete loss of expression in homozygous mice. When we produce mice from ES cell clones, we generate only heterozygous pups, and do not perform RT-PCR routinely to confirm the loss of gene expression. We would not trust the 50% reduction by qPCR in heterozygotes as a genuine confirmation. Instead, we QC the ES cell clone for the integrity of the gene trap and the correct location in the intron before we inject it.
Do you confirm that the targeted ES cells are null for expression of the targeted gene?
No, we don’t have capabilities to do this kind of analysis, but we can provide ES cell pellets if necessary. Keep in mind that ES cells are heterozygous, and it might be hard to tell the difference in expression of wild type vs. heterozygote. Moreover, a lot of genes are expressed in the ES cells at barely detectable levels. The best way to confirm a knockout is to obtain the homozygous mice. Past analysis based on 741 mice from the 129/SvEv resource has indicated that 96% of mice produced had no expression of the trapped gene, and the remaining mice were severe hypomorphs (Zambrowicz et al, 2003).
Do you confirm that the only insert site is in our gene of interest?
We perform quantitative PCR and sequence the product from an inverse PCR reaction to determine if the cell line of interest contains multiple gene trap inserts.
Is it possible that after investing to make a mouse knockout I discover that the homozygotes are embryonic lethal and the heterozygotes have sufficient protein so that there is no phenotype?
Yes, this is a possibility. However, you may find that your gene of interest may play an essential role during embryogenesis, and is worthy of study for a variety of reasons.
Could we obtain some DNA and RNA from the clone to perform a preliminary assessment?
We can only provide ES cell pellets and DNA samples for analysis. We can also provide a tail DNA sample from a confirmed heterozygous mouse upon request.
How do I test if my mice are indeed knockouts?
The best method to confirm mutagenesis in the gene trap lines is to perform an RNA-based analysis (such as RT-PCR, Taqman or Northern) on tissues known to express the gene once you obtain the homozygous mice. We can help you set up the assays necessary to make such a determination. Protein-based methods (Western, ELISA) may not be able to confirm knockout in the gene trapped lines.
